SECReTE10 domains in Aspergillus niger and Streptomyces lividans (Part 1)
- gerben voshol

- Oct 25, 2021
- 4 min read
SECReTE10 is a newly discovered mRNA domain, which plays an important role in secreted protein production. Recently, it has been shown that increasing the abundance of SECReTE10 domains can improve production and secretion of both heterologous and homologues proteins up to 30% (Cohen-Zontag et al., 2019). Below I present my own analysis based on this paper. For those who work on protein design and optimization, this information might be very helpful in finding new ways to increase protein production in various hosts. I use Aspergillus niger CBS513 and Streptomyces lividans 1326, two industrially relevant organisms, as an example.
What is SECReTE10?
SECReTE10 stands for secretion-enhancing cis regulatory targeting element. It is a stretch of pyrimidine (C and U) repeats every third base (NNY, NYN or YNN; where N = any nucleotide and Y = pyrimidine) that can be equal to or larger than 10 repeats in length. It was shown to aid in the localization of mRNAs
encoding secreted and membrane proteins (mSMPs) to the endoplasmic reticulum (ER). In line with this observation, the SECReTE10 domain was found to be enriched in mRNAs encoding secretome proteins translated on the ER in S. cerevisiae, S. pombe and H. sapiens.
SECReTE10 domains are enriched in secreted and transmembrane fractions of A. niger and S. lividans
After downloading the genomes of A. niger CBS513 and S. lividans 1326, I divided the coding sequences into a secreted (Sec), transmembrane (TM), cytopasmic (Cyt) and secretome (Sec + TM) fraction. For this, I used the translated nucleotide sequences and a hidden-Markov model trained on the signalP5.0 dataset. As a reference, I included the S. cerevisiae sequences and groupings of the original paper.
In the figure below (left), we can clearly see that both the secreted and transmembrane proteins contain approximately 1.25-1.6x as many SECReTE10 domains compared to the cytoplasmic fraction. Moreover, while the coding sequences in the cytoplasmic fraction contain 2.3 SECReTE10 domains on average, the secreted and transmembrane fractions contain 3.2 and 3.5 SECReTE10 domains on average, respectively (figure below right).

SECReTE10 abundance is largely explained by codon usage
In the original paper, the authors subsequently investigate what might be the cause of the SECReTE10 abundance in the different fractions. They investigate the effect of the presence and absence of transmembrane domains, removal of signal sequence coding regions, presence of other repetitive nucleotide triplets and codon usage. I decided to follow a similar approach and used permutation analysis and z-score calculations to determine the significance of the SECReTE10 domains. In addition to correcting for simple codon bias, I additionally included permutation analysis for CT bias, AA order bias and AA content bias (these last two with random codon usage). To control for the false discovery rate, a Benjamini-Hochberg threshold was used with a FDR value of 0.05. The results of the analysis are shown in the figures below.

As can be seen from the figure, correcting for CT bias in the sequence reduced the amount of significant SECReTE10 domains by approximately 20-60%. It can also be seen that the reduction is more or less equal for all four fractions, but is slightly higher (~10%) for the amount of significant cytoplasmic sequences. This might indicate that simple CT-bias can account for some of the SECReTE10 domains in the cytoplasmic fraction, but fails to explain the over-representation of SECReTE10 domains in the secretome fraction.
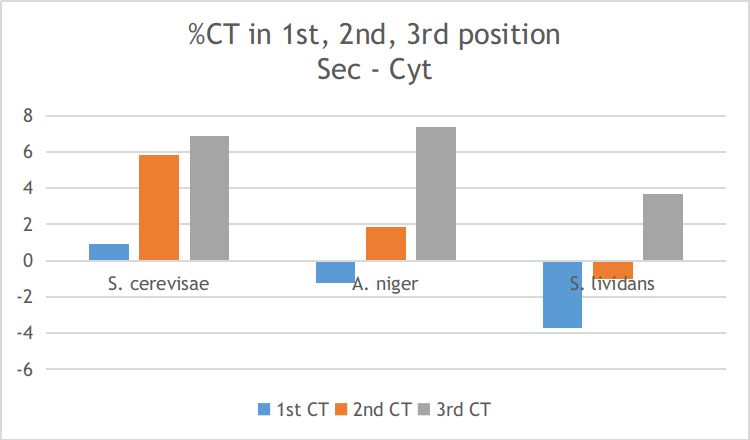
If however, we correct for codon bias, we can see that the abundance of sequences with a SECReTE10 domains in the secreted and cytoplasmic fraction is reduced and both these fractions reach a similar abundance level. The abundance of SECReTE10 domain containing sequences after correcting for codon bias is 0.05 secreted versus 0.1 cytoplasmic for S. cerevisae, 0.16 versus 0.14 for A. niger and 0.12 versus 0.03 for S. lividans. This indicates that codon usage is able to explain a large part of the overabundance of SECReTE10 containing sequences in the secreted fraction. This can also be seen in the figure below which shows that the secreted fraction has a higher abundance C or T (=U in mRNA) in the second or third position of its codons compared to the cytoplasmic sequences.
However, the SECReTE10 abundance in the transmembrane fraction is not reduced to the level of the cytoplasmic fraction by correcting for the codon bias. Therefore, I examined the SECReTE10 domains with a reduced alphabet hidden Markov model trained on the TMHMM dataset of those sequences which are significant after correcting for codon bias.
In A. niger, approximately 15% of the SECReTE10 domains completely overlap with predicted transmembrane helixes and another 36% show partial overlap.
A similar pattern is observed in S. lividans with 23% of the SECReTE10 domains overlapping transmembrane spanning regions and another 40% showing partial overlap with the beginning or end of the transmembrane domain. This suggests that for transmembrane sequences the SECReTE10 abundance is partially explained by the codon usage and by the AA order bias. Indeed, if we correct for AA order bias for those sequences that remain significantly enriched, the abundance of SECReTE10 domain containing sequences goes down significantly. The abundance is reduced to 0.07, 0.20 and 0.13 for S. cerevisae, A.niger and S. lividans, respectively, reaching levels similar tot the secreted and cytoplasmic fractions.
Summarizing the results, codon usage is one of the important factors explaining the overabundance of SECReTE10 domains in secreted and transmembrane fractions. However, that leaves us with several new questions such as: is codon usage caused by selection for SECReTE10 domains? Are the SECReTE10 domains caused by the codon usage? Is it a combination of both?
In my next blog post, I will use a simple codon optimizer in order to run Monte Carlo simulations to address these questions and dive deeper into the amino acid sequences that are preferentially encoded as SECReTE10 domains.



Comments